MITK
- Description of the platform/product:
- name and version of the software: MITK Workbench 2016.11 (not yet released)
- free? yes - http://mitk.org/Downloads
- commercial? no
- open source? yes - https://phabricator.mitk.org/source/mitk/
- what DICOM library do you use? DCMTK, GDCM, DCMQI
- Description of the relevant features of the platform:
- are both single and multiple segments supported? yes
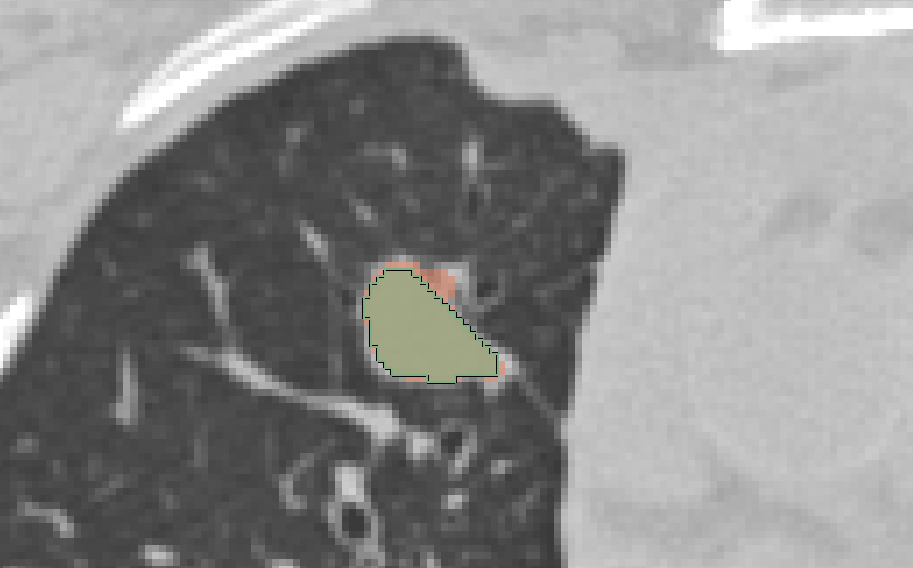
- how are the overlapping segments handled? With semi-transparent color overlay and the active label is contoured (see screenshots)
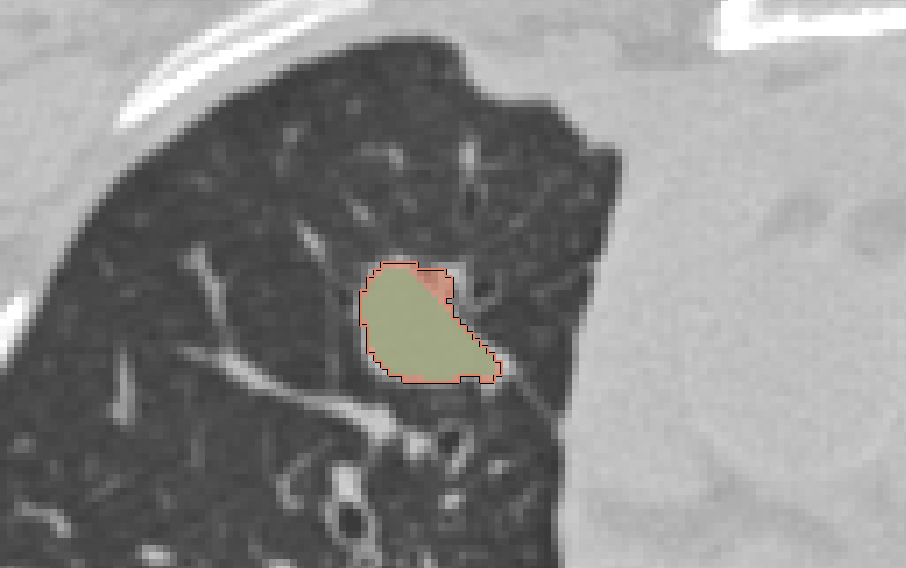 |
| 
- do you support both BINARY and FRACTIONAL segmentation types?
- do you render the segment using the color specified in the DICOM object? yes
- how do you communicate segment semantics to the user? semantic codes are stored as property for each label, so the user can not easily get the information
-
how do you support the user in defining the semantics of the object at the time segmentation is created? The user can select from a pre-defined list of organs, structures,... when adding a new label to the segmentation. The selection is mapped to segmentation category/type and color internally
-
Read task: load each of the DICOM SEG datasets that accompany the imaging series into your platform
Test dataset #1
| Test dataset | Result of rendering |
|---|---|
| 3D Slicer | 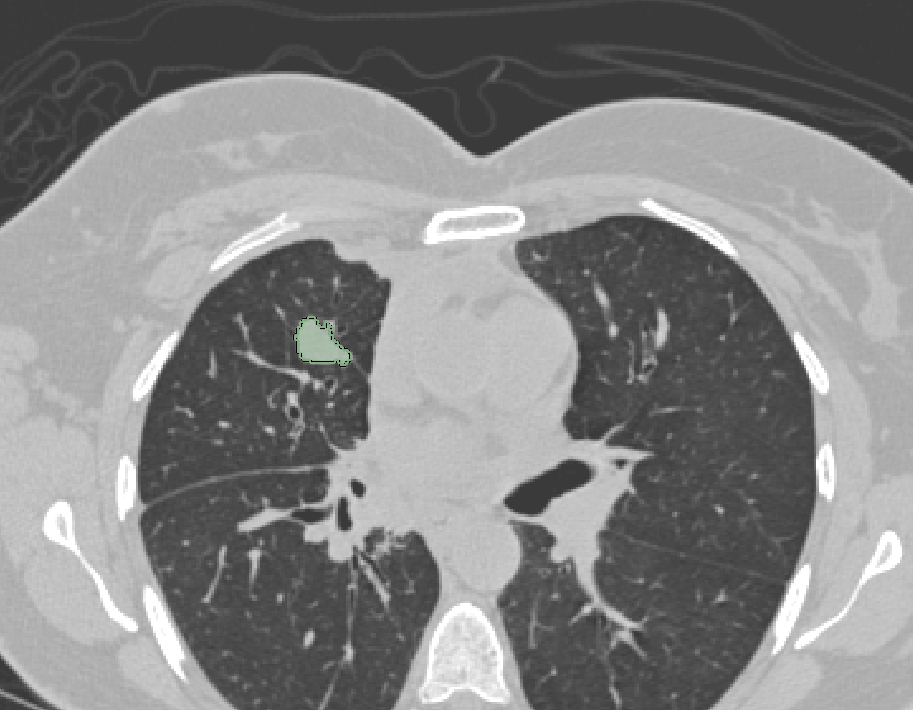 |
| AIMonClearCanvas | 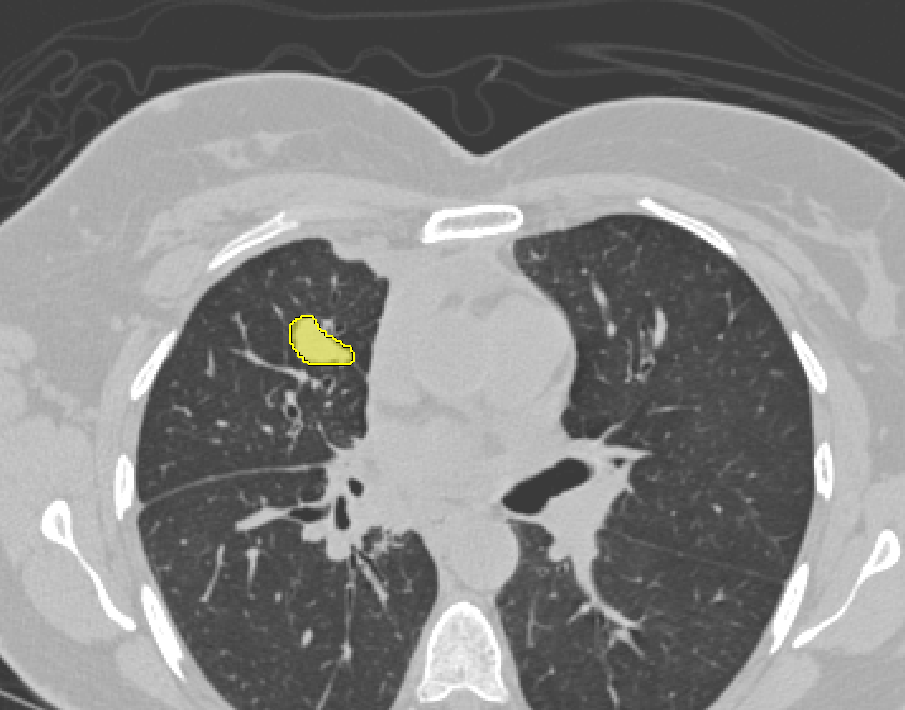 |
| ePAD | 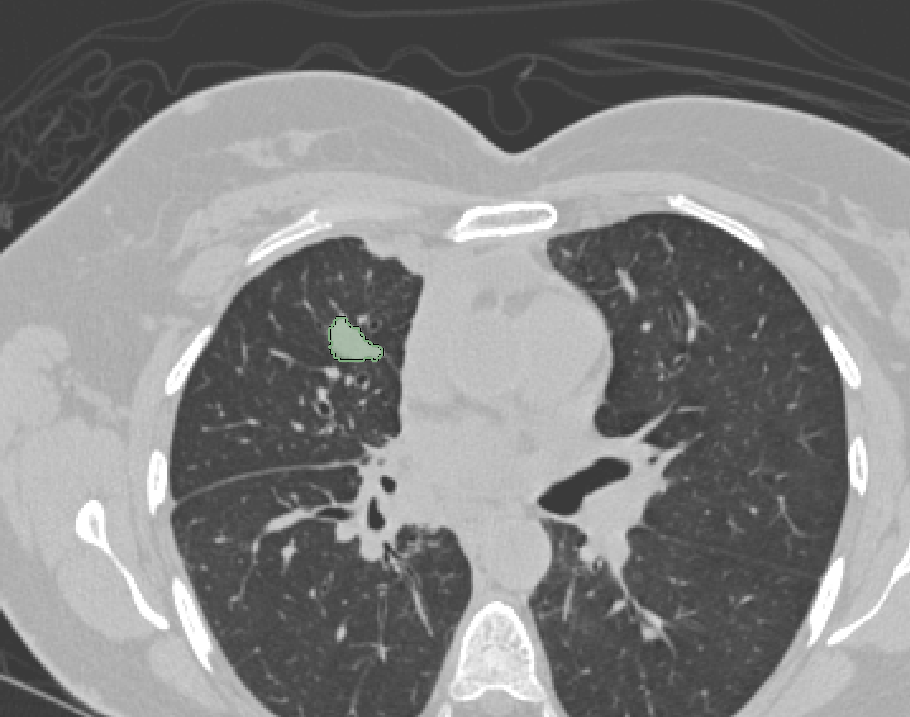 |
| syngo.via | 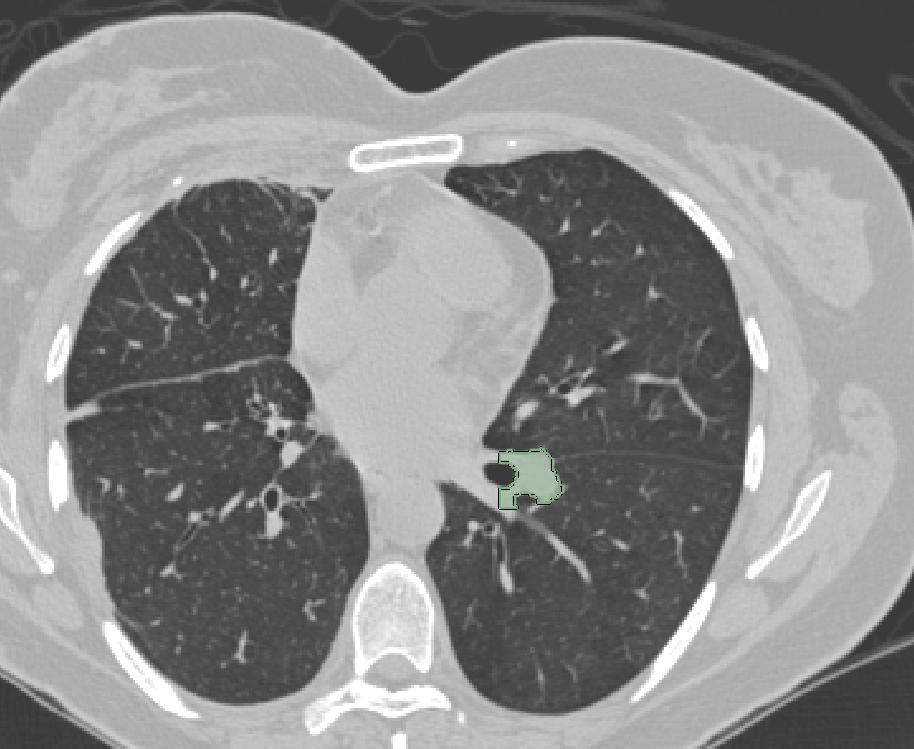 |
Test dataset #2
| Test dataset | Result of rendering |
|---|---|
| 3D Slicer | 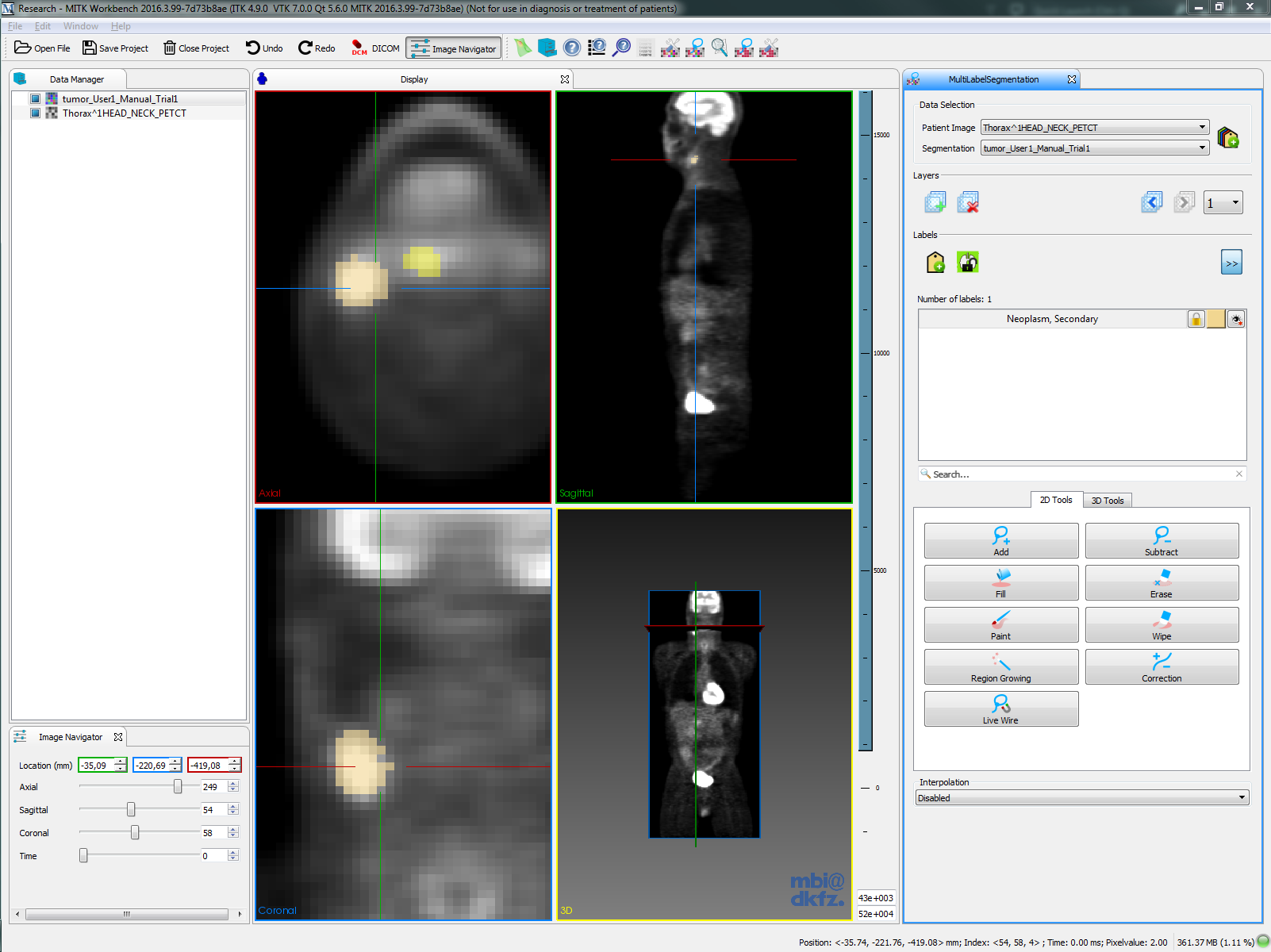 |
Test dataset #3
| Test dataset | Result of rendering |
|---|---|
| 3D Slicer | 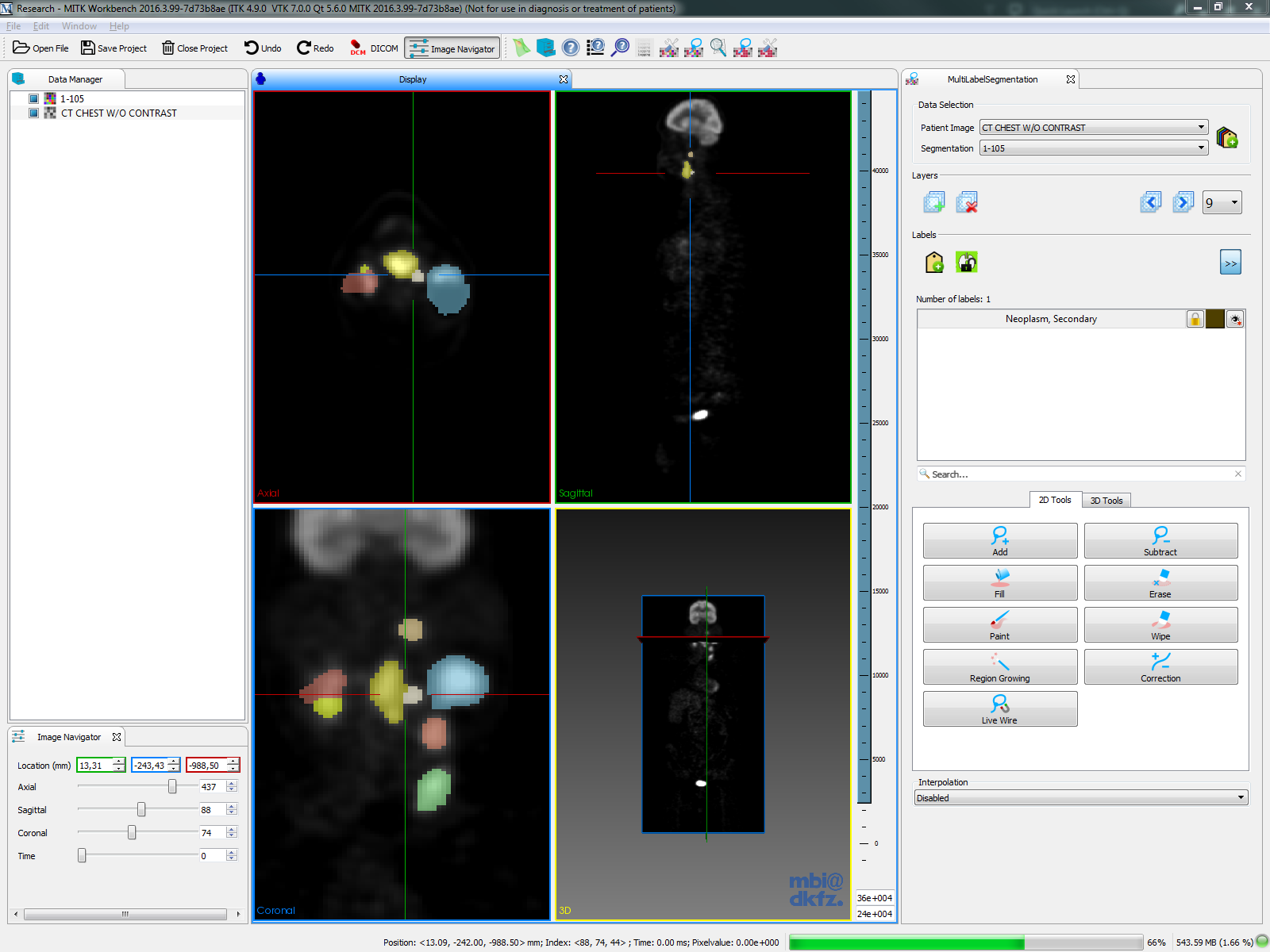 |
Test dataset #4
TODO:
| Test dataset | Result of rendering |
|---|---|
| 3D Slicer | 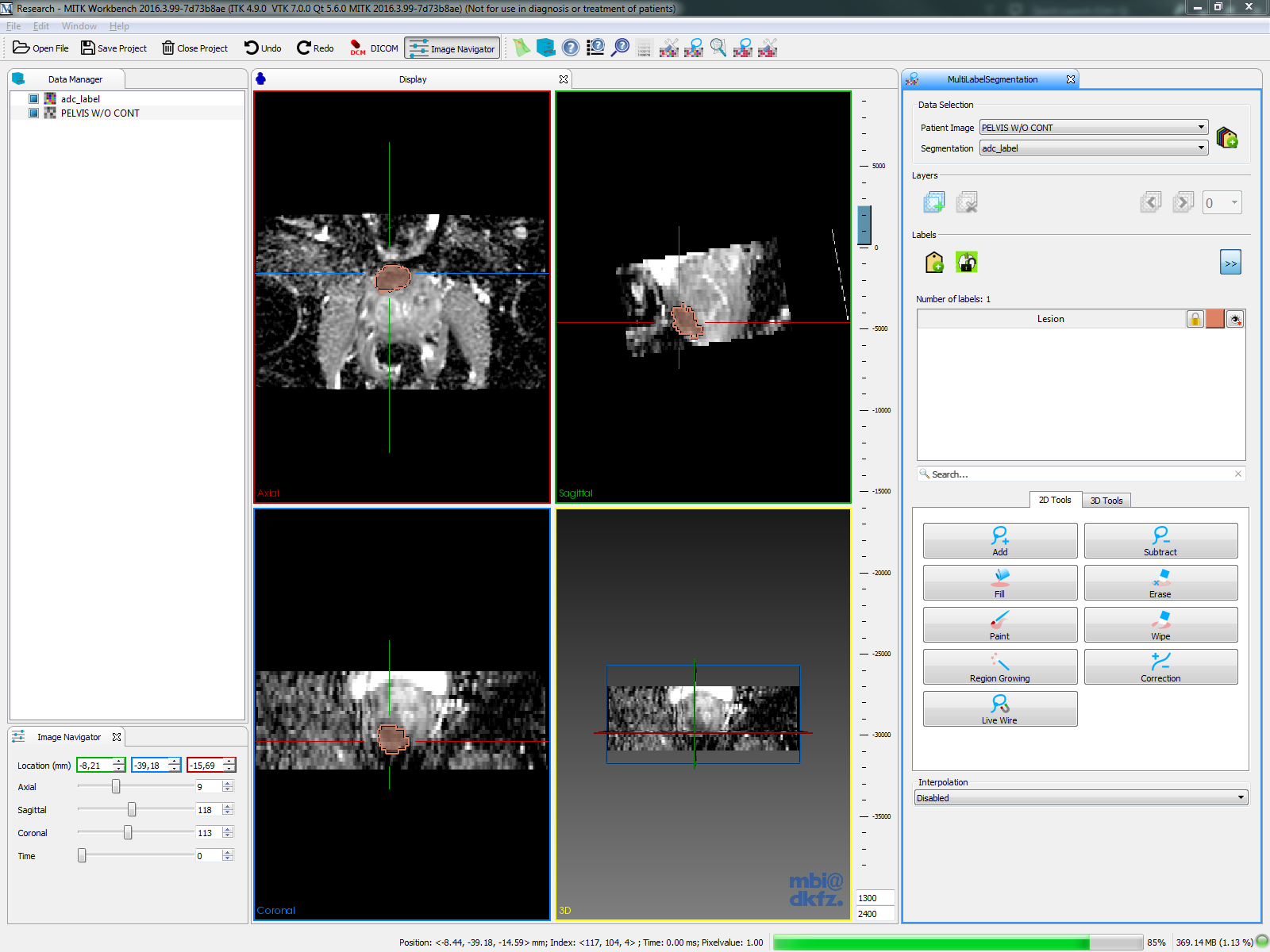 |
- Write task
- segment the lung lesion using any method available in your platform; save the result as DICOM SEG; please include in the series description the name of your tool to simplify comparison tasks!
- results are uploaded
- run dciodvfy DICOM validator; iterate on resolving the identified issues as necessary
- no errors, only warnings from dciodvfy