ePAD
1.Description of the platform/product:
- name and version of the software: ePAD, version 2.9
- free? yes https://epad.stanford.edu/epad-agreement
- commercial? no
- open source? yes except plugins and UI project github.com/RubinLab/
- what DICOM library do you use? PixelMed, DCM4CHE
2.Description of the relevant features of the platform:
- are both single and multiple segments supported? the new viewer uses AMI and supports multisegments; whereas the old viewer supports only single segments.
- how are the overlapping segments handled? user can select either to view the outline or as filled (see screenshot below showing both AIM on ClearCanvas and Slicer datasets from the Read task)
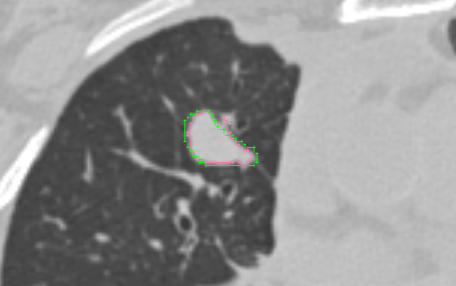
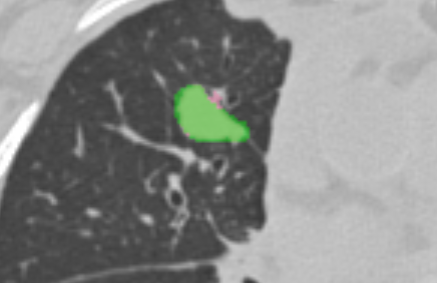
- do you support both BINARY and FRACTIONAL segmentation types? yes, segmentations that are saved as FRACTIONAL are mapped to gray scale (see screenshot below showing a FRACTIONAL segmentation)

- do you render the segment using the color specified in the DICOM object? the new viewer uses AMI and renders using the color specified in the DICOM object; whereas the old viewer uses annotation owner's preferred color for all segmentations
- how do you communicate segment semantics to the user? currently user has no means to get information about the semantics of the object as defined in the segmentation
- how do you support the user in defining the semantics of the object at the time segmentation is created? user is presented with a predefined list (same list with Slicer) to identify the semantics of the segmentation. the default values for category and type are both
(T-D0050;SRT;Tissue).
3.Read task: load each of the DICOM SEG datasets that accompany the imaging series into your platform
Test dataset #1
| Test dataset | Result of rendering |
|---|---|
| 3D Slicer | 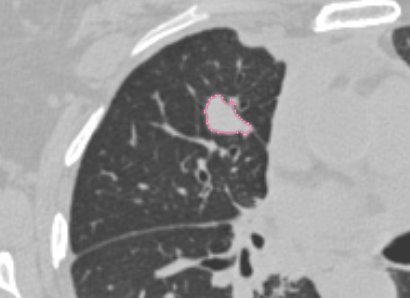 |
| ePAD | 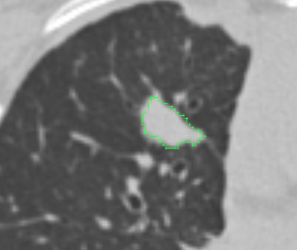 |
| syngo.via | 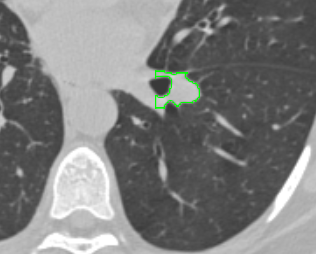 |
| AIMonClearCanvas | 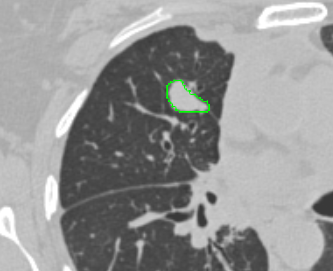 |
| Brainlab |  |
Test dataset #2
At this time, ePAD does not support multi-segment segmentations.
4.Write task
- segment the lung lesion using any method available in your platform; save the result as DICOM SEG; please include in the series description the name of your tool to simplify comparison tasks!
- results are uploaded
- run dciodvfy DICOM validator; iterate on resolving the identified issues as necessary
- no errors, only warnings from dciodvfy